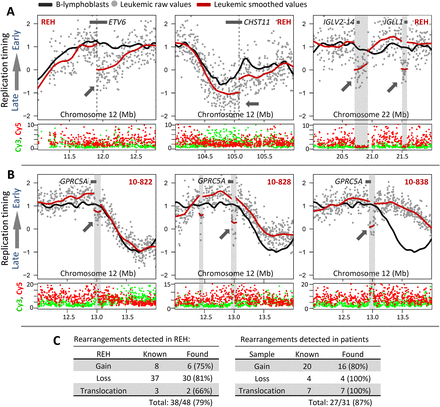
Abrupt shifts in replication-timing localize a subset of rearrangements. (A, left) Abrupt timing changes in REH at 12p13 map within ETV6 (TEL1) at 11.95 Mb, consistent with the molecularly mapped translocation site. (Middle) A breakpoint at 12q23 (94.8–107.5 Mb) can be mapped more precisely within CHST11 by an abrupt shift in timing values at 105.08 Mb. (Right) Abrupt timing changes in regions not included in published karyotypes of REH represent deletions of IGL loci involved in B-cell maturation, evidenced by a sharp drop in overall Cy5 and Cy3 signal intensity. (B) Examples of abrupt timing changes in patients 10-822, 10-828, and 10-838 undetected by karyotype analysis, but suggesting a shared amplification of ∼12.95–13.05 Mb in these three patients, which overlaps suspected tumor suppressor GPRC5A. (C) Summary of rearrangements detected in REH and patient samples (Supplemental Figs. S4–S8) by either CNV in raw replication-timing data or abrupt timing shifts lacking CNV (translocations).











