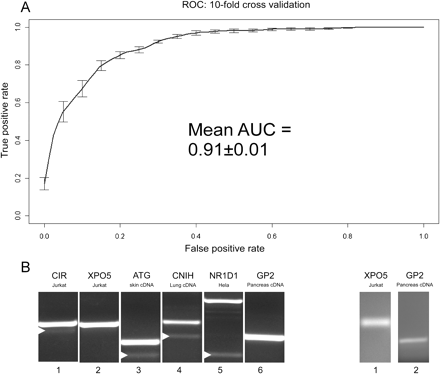
Accuracy of the machine-learning algorithm trained to classify exons as either constitutive or alternative and identification of novel alternatively spliced exons. (A) Average ROC and AUC values based on 10 cross-validation runs. (B) Identification of the skipping forms of six constitutive exons (100% inclusion, based on ESTs) that were identified as alternative using machine-learning algorithms. Splicing pattern validation using primers for flanking exons identified four exons (left panel, lanes 1,3,4,5). Validation using primers to junctions of flanking exons revealed skipped forms for two more exons (right panel).











