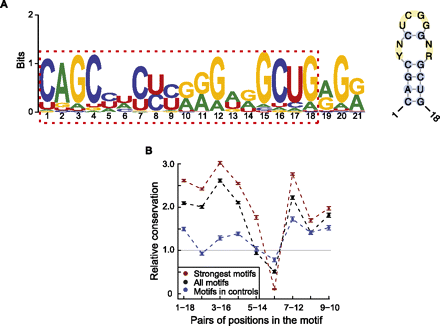
A novel motif with potential function in A-to-I editing. (A) Consensus motif (left) identified by MEME in the 201-nt neighborhood centered around each predicted A-to-I editing sites. (Right) Structure of the one to 18 bases of the consensus motif (RNAalifold). Y = U or C, R = G or A, N = A, C, G or U. (B) Conservation of the base-pairing patterns of the motif in primates based on multiz46way alignments. Strong motifs, motif score >24.4; all motifs, motif score >6.6; controls (motif score >6.6) were randomly picked from Alu elements in coding exons devoid of A-to-I editing sites. Error bars represent 95% confidence intervals. The conservation levels were normalized against expected levels calculated using random controls (Supplemental Methods).











