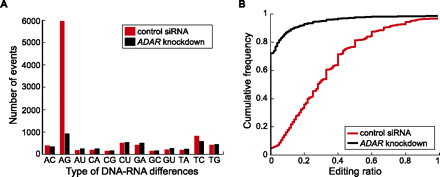
DNA–RNA differences identified via RNA-seq. (A) Number of events for the 12 types of differences between RNA reads and genomic DNA sequences in samples transfected with control siRNA and ADAR siRNA, respectively. Labels of x-axis denote DNA and RNA nucleotides (e.g.: “AC” denotes “A” in DNA and “C” in RNA). (B) Empirical cumulative distribution function of editing ratios of putative A-to-I editing events identified from RNA-seq. A union of editing events identified in the two samples is included (6422 in total) in each curve. For nonediting events in one sample (those that failed the statistical identification procedure), the editing ratio was calculated as the number of reads with the “G” nucleotide at the predicted editing position divided by the total number of reads at that position.











