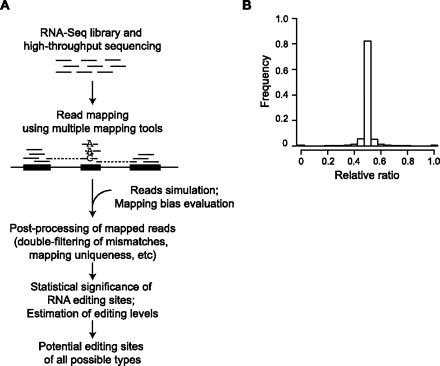
Identification of RNA editing sites. (A) Generative process of the pipeline. (B) Evaluation of mapping bias using simulated data. Histogram shows the distribution of relative ratios of all simulated genomic sites with alternative alleles. Relative ratio is defined as follows: (N_mapped_ref/N_simulated_ref)/(N_mapped_ref/N_simulated_ref + N_mapped_edit/N_simulated_edit), where N_mapped_ref is the number of reads mapped to the reference base (e.g., A for A-to-I editing) and N_mapped_edit is the number of reads mapped to the edited base. N_simulated_ref and N_simulated_edit are defined similarly, but for the originally simulated reads. The average of all relative ratios is 0.499 and median is 0.500, neither of which is significantly different from the expected ratio 0.5 (P = 0.1, P = 0.3, respectively).











