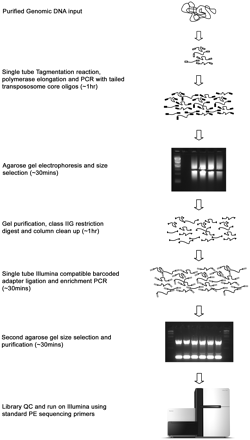
Schematic overview of modified Tagmentation procedure. Purified genomic DNA is tagmented using a specific ratio of enzyme and target (see Methods). Following tagmentation (addition of standard TRM oligo shown as white box), the reaction is quenched through the addition of premixed PCR reagents, subject to a brief extension step, and amplified with limited PCR cycles using a single tailed-oligo (black boxes) resulting in a library of fragments flanked by identical 29-bp sequences that can be size-selected by standard gel electrophoresis techniques. Tailed oligos contain a recognition site for a remote cutting type IIG restriction endonuclease that is used to remove the majority of the 29-bp flanking sequence, including the core transposon motif, and leaves a mandatory 2-bp 3′-TG overhang at both ends of all amplified library fragments. Modified Illumina sequencing adapters (gray boxes), incorporating variable length in-line bar-coding sequences, are then ligated to this 2-bp overhang in a highly efficient reaction. A second limited cycle PCR is performed directly on the ligation reaction and products run on an agarose gel and subject to a second round of size selection. The purified product is then sequenced on an Illumina GAII sequencer using the manufacturer's standard methods.











