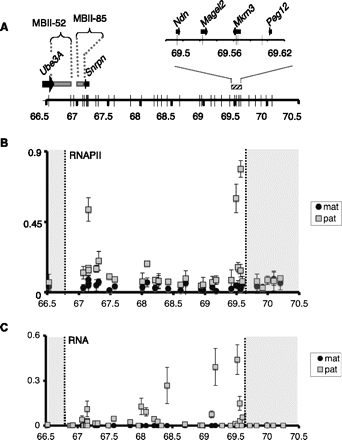
H3K36me3 deposition onto the silenced, maternally contributed Snurf–Snrpn region is independent of transcription. (A) Representation of the 3-Mb Snurf–Snrpn region as in Figure 1A. (B) ChIP analysis of RNAPII binding to the Snurf–Snrpn region, using brain chromatin from fetuses conceived by mating JF1/Ms males with C57BL/6 females. Strong enrichment can be observed at positions 67,149,376; 69,493,157; and 69,565,295 that corresponds to the first intron of Snrpn and to Ndn and Mrkrn3 promoters, respectively. Values are mean ± SD of two independent ChIP experiments. (C) RT-qPCR analysis of RNAs extracted from the brains of E17 fetuses conceived by crossing JF1/Ms males with C57BL/6 females. Relative levels of paternally (gray squares) and maternally (black circles) expressed RNAs were quantified using the set of 35 allele-specific primer pairs and normalized to Hprt mRNA. This assay identifies Snrpn, Magel2, and Mkrn3 mRNAs, but not Ndn mRNA due to the absence of suitable polymorphisms inside the transcript. Values are mean ± SD (n = 3 independent experiments).











