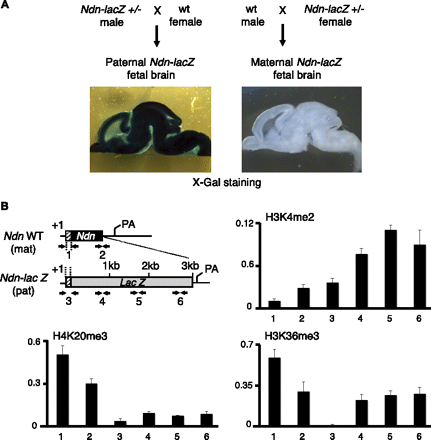
H3K36me3 is distributed differentially on the maternally and paternally contributed Ndn alleles. (A) Brain sections prepared from E17 fetuses heterozygous for the Ndn-lacZ allele were stained with X-gal, showing robust transcription of the paternally contributed Ndn-lacZ allele and strict transcriptional silencing of the maternally contributed Ndn-lacZ allele. (B) Structure of the single exon Ndn gene, as well as the Ndn-lacZ allele in which lacZ is inserted as a fusion with the first 31 Ndn codons (the remaining of Ndn coding sequence is deleted). Mice were bred to obtain fetuses carrying the wild-type (wt) Ndn and Ndn-lacZ fusion alleles on the maternally and paternally contributed chromosomes, respectively. Arrows indicate the positions of the PCR primers used in the qPCR assay. Primer pairs 1 and 2 recognize the promoter and 3′ end of the silenced, maternally contributed Ndn allele, respectively. Primer pair 3 maps to the promoter of the actively transcribed, paternally contributed Ndn-lacZ allele, and primer pairs 4, 5, and 6 to the lacZ reporter gene body. (+1) The transcription initiation site; (pA) the mRNA polyadenylation signal. Enrichment of H3K36me3, H3K4me2, and H4K20me3 was determined by ChIP using chromatin prepared from E17 fetal brains carrying a paternally contributed (transcribed) Ndn-lacZ, and a maternally contributed (silenced) wild-type Ndn allele. For each ChIP assay, the antibody-bound fraction was amplified by qPCR with the indicated primer pairs. Histone modification relative abundance is normalized to input. Values are mean ± SD (n = 3 independent ChIP experiments).











