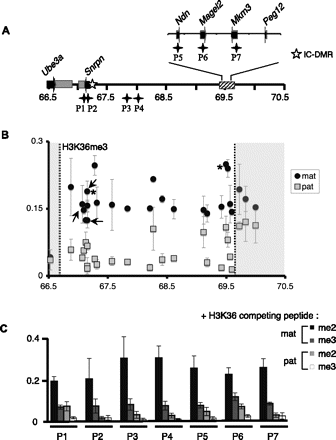
H3K36me3 is broadly distributed within the silenced, maternally inherited Snurf–Snrpn region. (A) Representation of the 3-Mb Snurf–Snrpn region is shown as in Figure 1A. Black stars indicate the location of regions P1 to P7 that were analyzed in the competition assay. P1 and P2 primer sets are amplifying DNA fragments within the IC-DMR. (B) H3K36me3 distribution within the Snurf–Snrpn domain. ChIP experiments were performed as described in Figure 1. The statistical significance of the data was verified for two positions indicated by asterisks (P < 5.10−5; Student's t-test realized with eight independent experiments). (C) The specificity of H3K36me3 distribution within the Snurf–Snrpn region was confirmed by competition assay. Chromatin and antibodies against H3K36me3 were co-incubated with H3 peptides di- or trimethylated at lysine 36. qPCR assays were performed using seven discriminating primer pairs, which amplify loci located in or at the vicinity of coding genes (P1, P2, P5–P7), or in non-coding regions (P3 and P4) (see Supplemental Table S1). Values are mean ± SD of two independent ChIP experiments.











