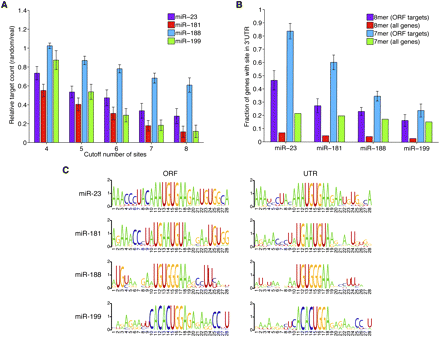
Impact of evolutionary processes on microRNA targeting of C2H2 genes. (A) Increased targeting due to intragenic C2H2 domain similarity. Shown are ratios giving the numbers of genes containing the indicated minimum number of 8mers for codon-randomized genes divided by the number of genes containing that many 8mers for real gene sequences. Values shown are the mean ratios across 50 codon-randomization trials. Error bars show standard deviation across the trials. All but three values were statistically significant (genes with four sites for miR-23, p < 0.01; genes with four or five sites for miR-188, p > 0.05; genes with four sites for miR-199, p > 0.05; all other sets, p < 10−4; paired Student's t-test with Bonferroni correction). (B) Propensity of ORF target genes to also contain 3′ UTR target sites. Shown are fractions of genes containing either 8mer or 7mer sites for the indicated miRNA within their 3′ UTRs, comparing the ORF target sets with the background of all genes. Error bars show standard deviations from 100 bootstrapping trials (p < 10−7 for all comparisons except miR-199 7mers; binomial test). (C) Evidence that 3′ UTR sites derived from ORF sites. Shown are nucleotide compositions flanking miRNA sites in both ORFs and 3′ UTRs of mRNAs with ORF sites. Letter size indicates nucleotide enrichment, visualized using WebLogo.











