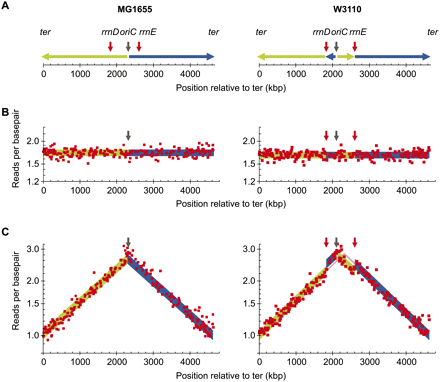
Simulation of detection of errors in de novo assembled bacterial sequences. (A) Chromosomal DNA is replicated bidirectionally from oriC (black arrow) indicated by the green line (left arm) and blue line (right arm). Strain W3110 (right) carries an inversion between the ribosomal operons rrnD and rrnE (red arrows) but is otherwise very similar to MG1665 (left). This inversion includes the origin of replication, oriC, and displaces oriC 215 kb to the left compared to MG1655. (Green and blue) The inverted chromosomal arms. Reads generated from DNA template of a stationary phase (B) and an exponentially growing culture of MG1655 (C) were mapped to the genomic sequence of MG1655 (left) and W3110 (right). The number of reads starting at each base pair was calculated for a 10-kb-wide sliding window (red squares), and windows covering repeat sequence were removed. The ideal read frequencies were calculated using an oriC/ter ratio of 2.7 and shown with the same color code as in A.











