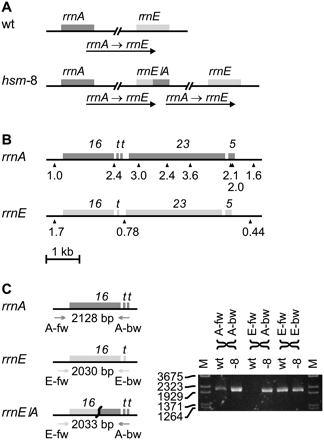
The rrnA–rrnE region of the chromosome of hsm-8 is duplicated. The read frequency analysis of hsm-8 (Fig. 1D) indicates a duplication of the rrnA–rrnE region. This duplication can be caused by homologous recombination between sequences repeated in the rrnA and rrnE ribosomal RNA operons creating a rrnE/A chimeric operon and duplicating the entire sequence between rrnA and rrnE. (A) The duplication in hsm-8 compared to wild type (wt). (B) A detailed read frequency analysis was made to narrow down the recombination point. (16) rrs genes for 16S RNA; (t) genes for tRNA; (23) rrl genes for 23S RNA; (5) rrf genes for 5S RNA. The read frequency of a 500-bp window on each side of the rrnA and rrnE operons and at positions with unique sequences (shown by triangles) within the rrn operons in hsm-8 was normalized to the read frequencies of the same positions in wt. The relative read frequency of the 500-bp window to the left of rrnA was set to 1. The read frequency shifts from 1.0 to higher than 2 on the opposite side of the 16S RNA gene. This indicates that the entire part of the rrnA operon downstream from the 16S RNA gene is duplicated. In the rrnE operon the read frequency drops fourfold from the left side to the right side of the operon. This fourfold drop is in agreement with Figure 1D and indicates a long DNA replication time for the rrnE operon. The read frequency at the only measurable point in the rrnE operon is lower than half the read frequency of the left side and higher than the read frequency of the right side and is less conclusive. The combined analysis of rrnA and rrnE indicates that recombination took place within the 16S RNA genes. (C, left) PCR primers were designed to amplify the rrsA, rrsE, and the rrsE/A chimeric genes. (Heads of the arrows) The approximate locations of the primers and the expected PCR product sizes are indicated. (C, right) PCR products obtained from MG1655 (wt) and from hsm-8 (-8). hsm-8 shows the native rrnA and rrnE operons as well as the rrnE/A chimeric operon, whereas MG1655 only shows the native rrnA and rrnE operons.











