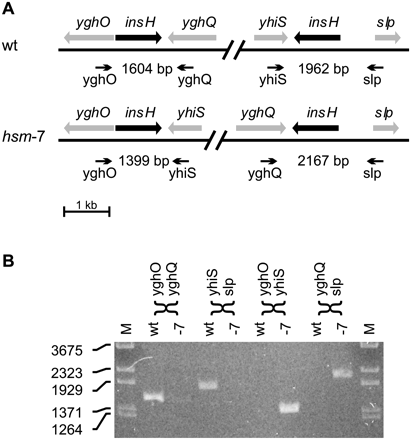
The chromosome of hsm-7 is inverted between two IS5 elements. The read frequency analysis of hsm-7 (Fig. 1C) indicates an inversion between two inverted copies of the insH gene encoding the IS5 transposase and trans-activator. (A) Genetic map of the two copies of insH and their neighboring genes in the wild-type (wt) and in the suggested hsm-7 configuration. The heads of the arrows indicate the approximate locations of PCR primers designed to differentiate between the wt and the hsm-7 configuration by amplifying the insH genes with neighboring sequences. The expected size of each PCR product is indicated. (B) PCR analysis of MG1655 DNA (wt) and hsm-7 DNA (-7) with the primers shown in A. MG1655 shows the expected wild-type fragments of 1604 bp and 1962 bp. hsm-7 shows the 1399-bp and the 2167-bp fragment expected from the inversion.











