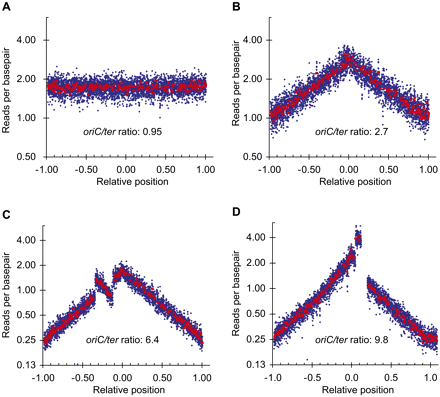
Chromosomal rearrangements detected by read frequency analysis. The number of reads starting at each base is plotted as a function of the relative position on the reference chromosome; oriC is set to 0, and ter is set to ±1 to reflect the bidirectional DNA replication. (Blue rhombus) The average of 1000-bp windows; (red squares) the average of 10.000-bp windows. Any window containing repeat sequences is omitted. The oriC/ter ratio is the ratio of reads per base pair at oriC to reads per base pair at ter. Sequencing template was isolated from: (A) an overnight culture of MG1655; (B) an exponentially growing culture of MG1655; (C) an exponentially growing culture of hsm-7; and (D) an exponentially growing culture of hsm-8. (C,D) Clear deviations from the pattern of the host strain MG1655 (B) indicating an inversion and a duplication, respectively.











