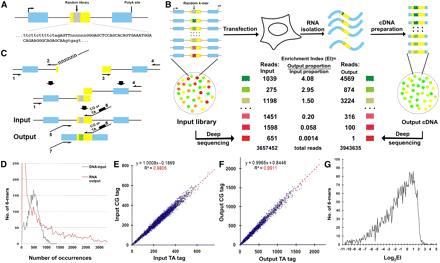
High-throughput definition of pre-mRNA splicing signals from sequence space. (A) The architecture of a linear minigene library. This minigene contains the Wilm's tumor gene 1 exon 5 (WT1-5) as a central exon (yellow box) flanked by sequences from a Dhfr minigene (blue boxes, Dhfr exons). A random 6-mer library (gray box) is located from +5 to +10 in the central exon (termed location WA) and the detailed sequences of the central exon and its 3′ and 5′ splice sites are shown. (B) The scheme of the high-throughput in vivo functional selection of splicing motifs. The Enrichment Index (EI) for a particular 6-mer is defined as the output proportion of this motif divided by its input proportion. The numbers represent six exemplary cases. (C) Minigene library construction. See Methods for details. (D) The distribution of the 4096 6-mers in the DNA input (dashed black) and RNA output (red) sequences. (E) Duplicate PCR preparations and sequencings of the DNA input library designated by the CG and TA barcodes yield very similar compositions. The proportions of the 6-mers are presented as counts per million reads. (F) The compositions of 6-mers in the RNA output sequences from two independent transfections labeled with either the CG or the TA barcode. The proportions of the 6-mers are presented as counts per million reads. (G) The distribution of 6-mer log2EI values. The results represent the average of two transfections.











