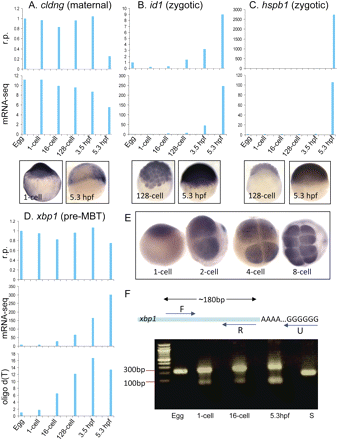
Validation of mRNA-seq expression clusters. (A–C) RT-qPCR of the maternal gene cldng and the zygotic genes id1 and hspb1 show similar expression patterns to those detected by mRNA-seq. WISH confirmed the expression pattern of these genes as observed in mRNA-seq. (D) RT-qPCR of the pre-MBT gene xbp1 showed a different expression profile from that of mRNA-seq when random primers (r.p.) were used, while it showed good correlation when oligo d(T) primers were used. (E) WISH of xbp1 demonstrates the presence of maternal transcript in the egg and the 1-cell stage and confirmed r.p. RT-qPCR data. (F) Measurement of xbp1 poly(A) tail length by RT-PCR in the egg and 1-cell, 16-cell, and 5.3-hpf stages. Total RNA was G/I-tailed and amplified with the xbp1-specific forward primer (F) and a universal oligo d(C) reverse primer (U). A control RT-PCR with xbp1-specific F and R primers gave a 180-bp fragment (S). The increasing poly(A) tail appears as a smear that increases gradually from the 1-cell stage to the 5.3-hpf stage, indicating the presence of an extended poly(A) tail.











