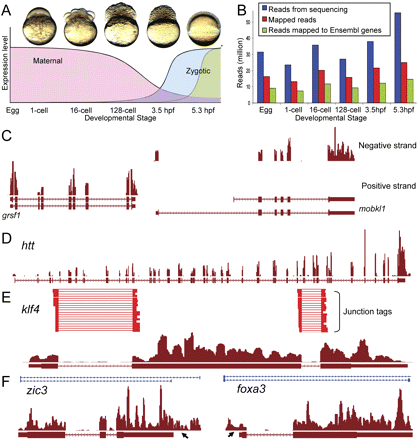
Overview of mRNA-seq data and mapping to the zebrafish genome. (A) Zebrafish embryonic developmental stages analyzed in our study. Libraries were generated from eggs, 1-cell, 16-cell, 128-cell, 3.5-hpf, and 5.3-hpf embryos. The graph depicts general expression patterns during these developmental stages. (B) Total number of mRNA-seq reads mapped in each developmental-stage library. Differences in the number of generated reads were observed, but the proportions of mapped reads were similar between all stages. (Blue bars) The number of reads generated from sequencing; (red bars) the number of reads mapped to the genome; (green bars) the number of reads mapped to Ensembl-annotated genes. (C–F) Mapping of mRNA-seq reads as viewed in the UCSC Genome Browser. Horizontal red bars at the bottom of each panel represent annotated exons (tallest boxes), UTRs (half-sized boxes), and introns (lines with arrowheads). (Arrowheads on bars) Orientation of transcript from 5′ to 3′. Mapped reads are represented by red histograms. (C) Strand-specific mapping in a region of chromosome 5, showing reads mapping to mobkl1aa in the negative, and grsf1 in the positive strands. (D) Reads mapping to the htt gene, containing 67 exons. Note the sharp exon–intron boundary of the mapping. (E) Mapping of exon–exon junction spanning reads (red horizontal lines) to klf4 locus, allowing the derivation of splice junctions. (F) Mapping of reads to regions beyond annotated 3′ (zic3) or 5′ UTRs (foxa3), indicated by arrows. Di-tags (blue horizontal bars at top of panel) provide support for this mapping.











