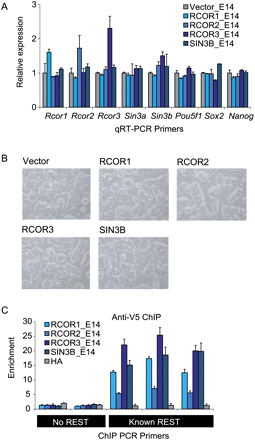
Low-level expression of tagged cofactor transgenes in mouse embryonic stem cells. (A) Quantitation of mRNA levels in ESC lines by qRT–PCR. cDNA samples from control (“Vector”) and corepressor-expressing cells (“RCOR1_E14” etc.) were interrogated using primers against corepressor mRNAs and pluripotency genes. Raw data were normalized to the housekeeping gene Actb, then displayed as a fold difference compared with Vector control. Error bars indicate the standard error of the mean for triplicate biological experiments. (B) Bright field microscopy of mouse embryonic stem cell lines used in this study. FACS-sorted and replated cells display normal, undifferentiated morphology. (C) Chromatin immunoprecipitations of V5-tagged cofactor proteins were carried out using anti-V5. ChIP DNA was interrogated in qPCR with primers for three known REST target loci (Johnson et al. 2008) as positive controls, and two loci with no evidence for REST recruitment as negative controls. A sham HA antibody was used to immunoprecipitate chromatin from the RCOR1_E14 line as a further control. Enrichments were calculated with reference to a non-REST background PCR amplicon. ChIP experiments were carried out on at least three independent cell cultures.











