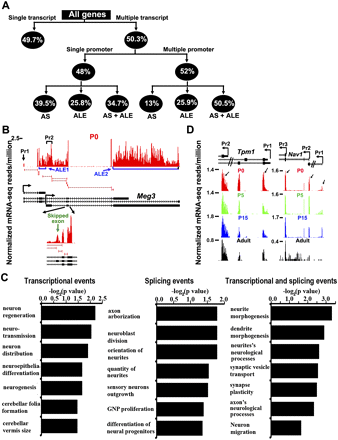
Alternative promoter usage and expression of promoters and transcripts in the four stages of postnatal cerebella. (A) The occurrence of alternative splicing (AS) and alternative last exon (ALE) events in both single-promoter and multipromoter-driven genes that express multiple transcripts in postnatal cerebellum. (B) Wiggle profile of mRNA-seq data for Meg3 shows the expression of multiple transcript variants through the use of alternative promoters/alternative first exon (AFE), ALE, and AS in P0. The black and blue arrows point to the coexpressed AFE and ALE, respectively. (Bottom) The zoomed in regions of exon skipping (ES) (green arrow) and supporting splice junctions in P0. (C) Ingenuity pathway analysis reveals the enriched functions (P < 0.05) among the genes that exhibit only transcriptional events, only splicing events, and both transcriptional and splicing events. (D) Wiggle profile for Tpm1 (left) and Nav1 (right) from mRNA-seq shows the expression of multiple promoters and transcripts in all stages or in a stage-restrictive manner, respectively. While both promoters are active for Tpm1, only promoter 3 for Nav1 is active in all four stages. Promoters 1 and 2 of Nav1 are development specific and not active in adult cerebella. The promoter-specific AFE expression is indicated by the black arrows. (Top) The pre-mRNAs for each gene and the corresponding promoter and TSS.











