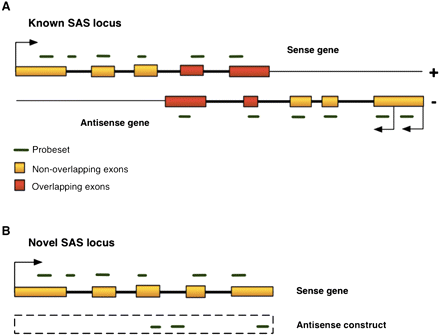
Probesets mapping to known and novel SAS genes. (A) Schematic diagram of a hypothetical known SAS gene pair shows the structural arrangement of overlapping exons (red rectangles) and nonoverlapping exons (orange rectangles). In our analysis, each partner gene is, in turn, treated as the antisense gene. Probesets (horizontal green dashes) map to the sense strand of the gene in either exons or introns. (B) A novel SAS sense gene. Since the structure of the transcribed antisense RNA is unknown, an antisense construct (dashed-line box) spans the genomic coordinates of the sense gene and approximates antisense expression. All antisense probesets encompassed by that region are used to infer the expression of the antisense construct. If the actual antisense transcript extends beyond the sense gene boundaries, the antisense construct expression under-represents the actual level of antisense transcription at that locus.











