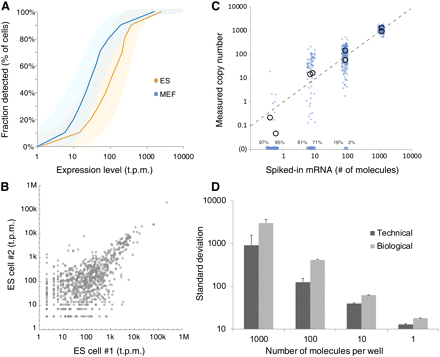
Quantitative accuracy. (A) Shows the probability of detection as a function of expression level for ES and MEF cells (shaded areas show 95% intervals). (B) Representative single-cell scatterplot showing the set of genes belonging to the top 1000 in ES and MEF cells (1465 in total). (C) The measured copy number for each of eight synthetic control mRNAs, across the entire plate. Circles show averages, whereas blue dots show individual data points (jittered for clarity). Zero measurements are shown along the horizontal axis, with percentage zeros indicated. For comparison, the dashed line indicates the ideal 45° slope. (D) Comparison of technical variance (based on control RNA) and biological variance (based on the average of genes with expression levels within ±20% of the indicated copy number) across the entire plate. Error bars, 95% confidence intervals; in each case the confidence intervals were nonoverlapping.











