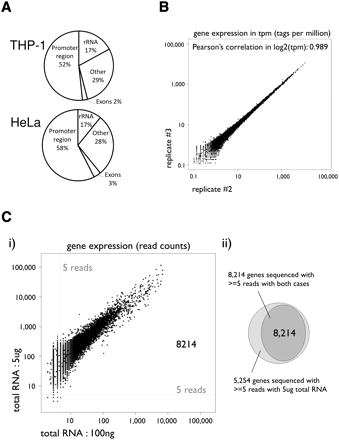
HeliScopeCAGE is a highly quantitative reproducible technology. (A) Average distribution of HeliScopeCAGE tags on annotated regions of the genome for the THP-1 and HeLa libraries. (B) Scatterplot of gene expressions between two technical replicates of HeliScopeCAGE on THP-1 RNA (5 μg of total RNA as starting material). The CAGE tag counts mapped within ±500 bp from the RefSeq transcription starting site are normalized as TPM (tags per million) with the library sizes. (C) (i) Gene expressions between different starting materials, 5 μg and 100 ng of total RNA of THP-1. Scatterplots of the two profiles with read counts. (ii) The number of detected genes with each profile. A gene is considered detected when five or more reads are obtained. Note: Given that mRNA is present at ∼1% of total RNA, A indicates a 300–500-fold enrichment of signal at promoters compared to rRNAs.











