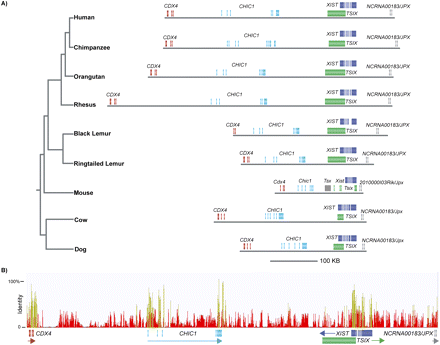
Candidate XIC genomic content. (A, left) The general phylogeny of the mammalian species included in this study (Murphy et al. 2007; Horvath et al. 2008; Perelman et al. 2011). Candidate XIC sequences were downloaded from UCSC for all species (except the lemurs). The lemur sequences were obtained from overlapping BACs and do not span the entire human candidate XIC region. Note that the ring-tailed lemur sequence begins at exon 1 of CDX4, while the black lemur sequence does not begin until exon 2 of the CDX4 gene. Both lemur sequences extend through the first two exons of NCRNA00183 (JPX/ENOX). All sequences were aligned using MLAGAN (see Methods). (Horizontal gray lines) Relative sizes of the candidate XIC in all species. (Colored boxes) DNA sequences orthologous to the annotated human gene structure. In dog and cow, the human TSIX sequence was used for annotation since there is no TSIX annotation in these species. The marmoset is not shown in this figure because the genome assembly is not complete across this region. Mouse 2010000l03Rik is now referred to as Enox. (B) Sequence identity plot from zero to 100% identity across the candidate XIC region for the multispecies alignment in A. (Yellow peaks) Regions of higher identity relative to red peaks or regions with no peaks. (Colored boxes) The relative positions of exons.











