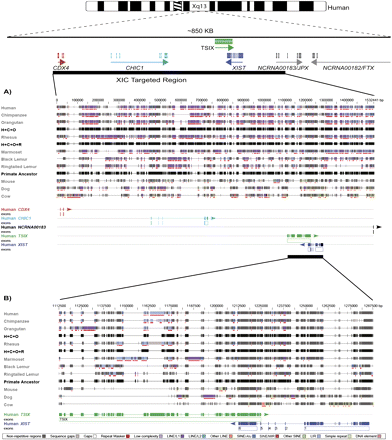
Repeat content and alignment across the XIC. (A) The candidate XIC region is shown along the top with a horizontal black bar indicating the smaller XIC region targeted in this study. Aligned sequence for each species is color-coded based on DNA sequence content, with nonrepetitive sequence indicated by black shading and different repeats color-coded according to the legend along the bottom. (Red bars below the sequence) Repetitive regions identified by RepeatMasker. Three ancestrally reconstructed sequences (H+C+O, H+C+O+R, and Primate Ancestor) are indicated for comparison of which regions have been gained or lost throughout primate evolution. Below the aligned sequences are the exonic regions annotated based on the human gene structures. The two exons of JPX occupy such a small space that they do not resolve as separate entities in this overview. (B) The aligned region expanded for TSIX and XIST is shown.











