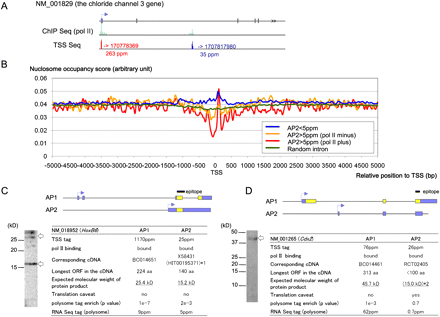
Characterization of the APs in DLD-1 cells via transcriptome data integration. (A) An example of APs for which both the TSS-seq and the ChIP-seq of pol II analyses supported simultaneous expression in a single gene in DLD-1 cells. (B) Nucleosome structures in the regions that surround the TSCs for second or later APs (indicated as “AP2”), which were expressed at <5 ppm (blue line), overlapped pol II binding sites (red line), or did not overlap pol II binding sites (yellow line). The nucleosome structures at the randomly selected intronic regions according to RefSeq information are also shown (green line). (C,D) Integration of transcriptome data and Western blotting for the HOXB6 (NM_018952; C) and CDX2 (NM_001265; D) genes. Bands of the expected molecular weights are indicated by arrows. Blue and yellow boxes represent predicted untranslated regions and CDSs, respectively. The peptides that were used to raise the antibodies are shown in the margin. (*1) The presence of multiple proteins was also suggested by UniProt (P17509 and P17509-2). (*2) The amino acid sequence had to be deduced from the cDNA sequence that overlapped with AP1, although this sequence lacked the canonical ATG initiator codon.











