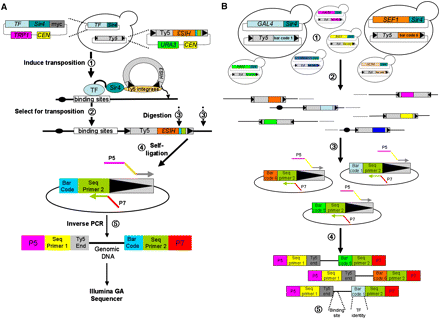
Calling Card–seq. (A) A DNA-binding protein fused to Sir4 directs integration of Ty5 into the genome near to where it binds. (1) After Ty5 transposition, (2) cells that have undergone Ty5 transposition are selected. (3) Genomic DNA is isolated and cleaved with restriction enzymes that cut near the end of Ty5 and (4) ligated in a dilute solution to favor recircularization of the fragments. (5) This is followed by amplification of the circular DNA that contains the end of the transposon and flanking genomic DNA by an “inverse PCR” (the PCR primers contain the Illumina sequencing primers and adaptors). The DNA sequence of the inverse-PCR products is then determined on an Illumina Genome Analyzer II. (B) Analyzing multiple TFs in one experiment: (1) Each strain was cotransformed with a plasmid encoding a TF–Sir4 fusion and a plasmid carrying its matched barcoded Ty5 Calling Card. (2) After transposition, the Calling Cards are deposited across the genome and then (3) recovered by inverse PCR and (4) sequenced on the Illumina GAII with a paired-end module. (5) For each paired sequence, we identify the Calling Card insertion site and the TF that deposited it there.











