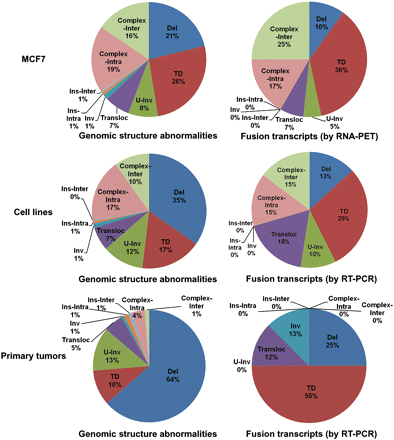
Different structure variation types seen in all genomic structure abnormalities and in only those giving rise to fusion transcripts in breast cancer genomes. Fusion transcripts detected by RNA-PET and validated by RT-PCR (top). Fusion transcripts (FGR + 3′T-ER) identified through the RT-PCR screening in three cell lines (middle) and five primary tumors (bottom). (Del) Deletion; (TD) tandem duplication; (U-Inv) unpaired-inversion; (Transloc) isolated translocation; (Inv) inversion; (Ins-Intra) intrachromosomal insertion; (Ins-Inter) interchromosomal insertion; (Complex-Intra and Complex-Inter) intra- and interchromosomal connections in hot spot of genome breakpoints (super cluster size ≥3) (Hillmer et al. 2011).











