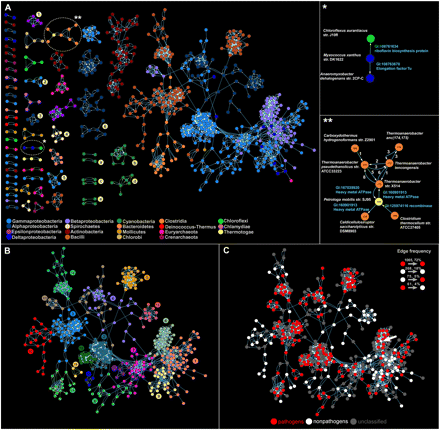
(A) The directed network of recent lateral gene transfers. Node color corresponds to the taxonomic group of donors and recipients listed at the bottom. Connected components of endosymbionts are marked with numbers: (1) Helicobacter, (2) Coxiella, (3) Bartonella, (4) Leptospira, (5) Legionella, (6) Ehrlichia. Clusters of cyanobacteria are marked with letters: (a) high-light adapted Prochlorococcus, (b) low-light adapted Prochlorococcus, (c) marine Synechococcus, (d) other Synechococcus, (e) Nostocales and Chroococcales. Enlarged images of clusters (right) are marked with asterisks. Species names are written by the vertices. Annotations of transferred genes appear next to the edges. (B) Community structure within the largest connected component of the dLGT network (for the entire network, see Supplemental Fig. S2). Vertices that are grouped into the same module are colored the same. (C) Pathogens in the largest connected component of the dLGT network (for the entire network, see Supplemental Fig. S6). The arrow marks a nonpathogen (Bukholderia thailandensis) within a pathogenic community.











