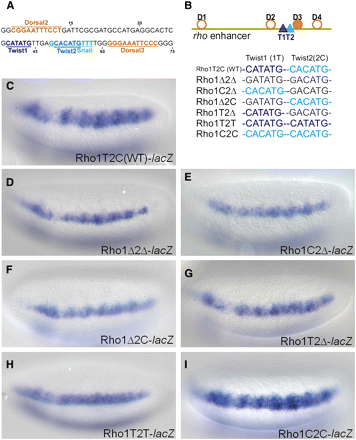
Mutagenesis of Twist binding sites at the ChIP-seq peak summit of rho enhancer. (A) The 75 bp sequence from the rho minimal enhancer which contains binding sites for Twist as well as for the transcription factors Dorsal and Snail. E-box sequences CATATG (T1, dark blue) and CACATG (T2, light blue) are separated by 5 bp, and Dorsal binding sites (orange) are positioned upstream and downstream of Twist sites. A Snail site that overlaps with T2 E-box is shown in green. (B) A diagram of the minimal 299 bp rho enhancer showing the relative positions of sites for Twist (dark and light blue triangles) and Dorsal (orange circles and filled circles, showing non-canonical and canonical sites, respectively). Lower schematic shows color-coded representations of the WT or mutant Twist binding sites present in various reporter constructs. Single nucleotide mutations were introduced into either T1 or T2 to eliminate binding (black: CATATG>GATATG or CACATG>GACATG) or to convert one site to the other (light blue: CATATG>CACATG or dark blue: CACATG>CATATG). (C) In situ staining of the wild type construct, minimal rho enhancer attached to the evep.lacZ reporter. (D) The Rho1Δ2Δ double mutant containing point mutations in both of the E-boxes, T1 and T2, supports reporter gene expression that is significantly weakened and more narrow compared to wild type (C). (E–G) Single mutations support expression that is weaker than wild type (C), more similar to the double mutant (D). (H) When a CATATG E-box is present in both the T1 and T2 positions, this change severely affects the expression domain of the reporter gene, reducing it to levels comparable to those observed in the double mutant Rho1Δ2Δ embryos (D). (I) When a CACATG E-box is present in both the T1 and T2 positions, the expression supported is comparable to the wild type (C).











