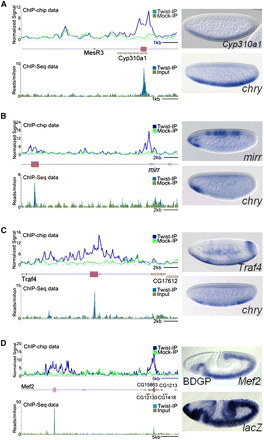
In vivo Twist occupancy supported by Twist ChIP-seq identifies functional CRMs. Representative examples of newly identified enhancers (brown boxes) and those previously identified (pink boxes) are shown for Cyp310a1 (A), mirr (B), Traf4 (C), and Mef2 (D). Upper left panels show ChIP-chip data and lower left panels show ChIP-seq data for Twist-IP and control samples. In upper right panels, lateral views of whole mount in situ hybridizations of the endogenous genes of stage 5–8 embryos are shown. In lower right panels, lateral views of whole mount in situ hybridizations of similar staged embryos containing either cherry (for Traf4, mirr, and Cyp310a1 enhancers) or lacZ (for Mef2 5′ enhancer) reporter constructs.











