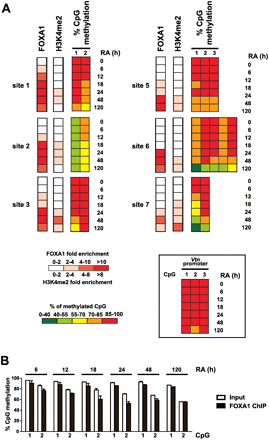
Kinetics of FOXA1 recruitment and epigenetic switch at enhancers during the course of P19 cell neural differentiation. (A) FOXA1 and H3K4me2 ChIP-qPCR as well as bisulfite-pyrosequencing experiments were performed in unstimulated or RA-treated P19 cells and analyzed as in Figure 4. For DNA methylation data, the percentage of cytosine methylation for CpG dinucleotides found within 200 bp of the center of FOXA1-binding sites is indicated. Note that significant DNA demethylation (p < 0.05) was observed for all analyzed CpGs except CpG 2 of site 3. On the other hand, no significant decreases in cytosine methylation levels were obtained at control CpGs localized within the Vtn promoter (inset). These CpGs had been previously identified as methylated in both undifferentiated and RA-stimulated P19 cells (A Sérandour and G Salbert, unpubl.). (B) Bisulfite-pyrosequencing was used to monitor the methylation status of CpGs found in site 1 within DNA issued from FOXA1 ChIP. Input DNA was used as a reference in these experiments. P19 cells were harvested and processed for ChIP assays after 6, 12, 18, 24, 48, or 120 h of RA treatment, as indicated. Results are means and standard deviations from two independent experiments performed in duplicates.











