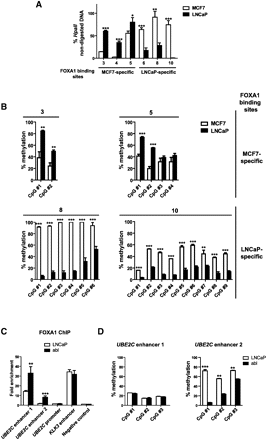
Differential 5-methylcytosine levels within CpGs present at cell-type-specific FOXA1-binding sites. (A) Genomic DNA from MCF7 or LNCaP cells were subjected to HpaII digestion. The percentage of non-digested DNA was determined by qPCR using primers allowing for amplification of a DNA fragment encompassing a CCGG motif localized within cell-type-specific FOXA1-binding sites. Numbering of FOXA1-binding sites is consistent with Figure 1E. Results are means and standard deviations from three independent experiments. (B) The percentage of cytosine methylation for CpG dinucleotides found within 200 bp of the center of cell-type-specific FOXA1-binding sites was determined using bisulfite pyrosequencing. Two MCF7- and two LNCaP-specific sites were analyzed as described in the Methods section. Numbering of FOXA1-binding sites is consistent with Figure 1E. Results are means and standard deviations from a representative experiment performed in triplicates. (C) FOXA1 ChIP-qPCR experiments were performed in LNCaP and abl cells. Fold enrichments relative to a negative control region are indicated. Results are means and standards deviations from three independent experiments. (D) The percentage of cytosine methylation within CpG dinucleotides found near the center of UBE2C enhancers 1 and 2 was determined using bisulfite pyrosequencing in LNCaP and abl cells as indicated. Results are means and standard deviations from a representative experiment performed in triplicates.











