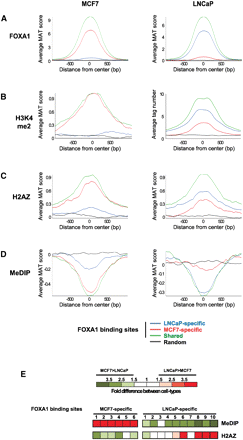
Differential H2AFZ and DNA methylation levels are linked to cell-type-specific FOXA1 recruitment to chromatin. Average FOXA1 (A), H3K4me2 (B), H2AFZ (C), and DNA methylation (D) enrichment levels at MCF7-specific, LNCaP-specific, or shared FOXA1-binding sites. Average signals were determined from tiling array data obtained in MCF7 and LNCaP cells as described in the Methods section except for H3K4me2 data in LNCaP cells, which were obtained from the ChIP-seq data of He et al. (2010). A random set of regions from chromosomes 8, 11, and 12 was also analyzed. (E) MeDIP-qPCR and H2AFZ ChIP-qPCR were performed in both MCF7 and LNCaP cells. For each analyzed FOXA1-binding site, the strongest enrichment was divided by the one obtained in the other cell type. A color code was used to show fold differences between the two cell types for both MeDIP and H2AFZ levels. Fold differences were inferred from data obtained in three independent experiments.











