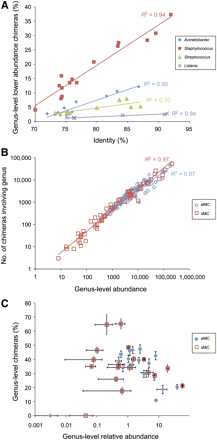
Correlation of chimera content with sequence homology and organism abundance. (A) Percent of other organism abundance corresponding to chimeras with the indicated more abundant species (y-axis), plotted according to percent identity (x-axis) between homologous 16S genes. (B) Number of chimeric sequences corresponding to a given genus were plotted as a function of total genus-level classified reads for the even (eMC) and staggered (sMC) mock community. Total read counts were based on best BLASTN match (E ≤ 10−10) to reference sequences for nonchimeras in addition to the genus representation within the CS-predicted chimeras. (C) Percent of sequences that correspond to chimeras for each genus plotted according to genus-level sequence abundance. Error bars correspond to standard error from the mean based on four technical replicates.











