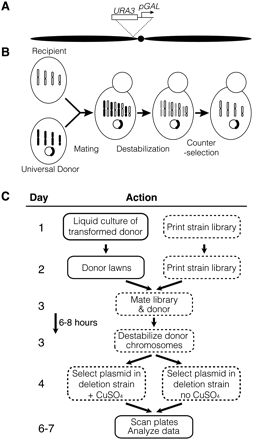
Selective ploidy ablation (SPA) mechanism and protocol. (A) Selective ploidy ablation relies on conditionally stable chromosomes. The strong galactose-inducible promoter (arrow labeled “pGAL”) was inserted adjacent to the centromere (black circle) on each of the 16 yeast chromosomes (black horizontal ovals representing two chromosome arms). The URA3 gene from Kluyveromyces lactis (open box) serves as a counter-selectable genetic marker to ensure loss of each chromosome. (B) Mating-based plasmid transfer is achieved using a universal donor containing 16 conditionally stable chromosomes (four black bars), which is transformed with an assay plasmid (circle). The recipient strain (oval with open bars for unmodified chromosomes) mates to the universal donor to make a strain heterozygous for conditionally stable and wild-type chromosomes (mix of black and open bars). Growth on galactose destabilizes the URA3-marked chromosomes so that they are lost during mitotic growth (gray bars), while the wild-type chromosomes are maintained (open bars). Counter-selection against URA3 ensures that all conditionally stable chromosomes are lost while the plasmid is maintained in the new strain. (C) Flow chart illustrating the SPA screen method. Transformation of a plasmid into the universal donor (data not shown) can be performed days or weeks prior to the SPA method. Replica pinning steps are represented as boxes with dashed lines. CuSO4 listed at step 4 is used to induce expression from the plasmid.











