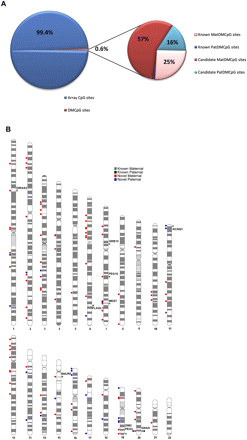
Distribution of known and candidate DMCpG sites. (A) Pie chart showing the frequency of the identified (∼0.6%) DMCpG sites with the smaller pie chart illustrating the frequency of only the known and the candidate DMCpG sites. (B) Chromosome ideogram showing the distribution across all autosomes of known (chromosome ideogram, right) and candidate (chromosome ideogram, left) DMCpG sites. Green and black dots correspond to known maternally and paternally methylated CpG sites, respectively. Red and blue dots correspond to candidate maternally and paternally methylated CpG sites, respectively. The names of imprinted genes are displayed next to their associated known DMCpG sites.











