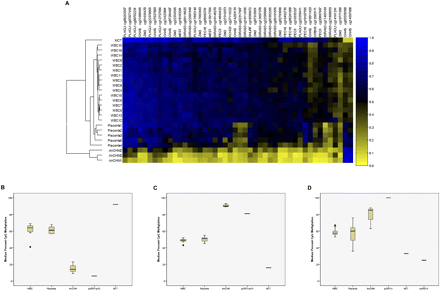
CpG methylation in differentially methylated regions (DMRs). (A) Heatmap showing the methylation levels of CpG sites in DMRs of known imprinted genes. The rows represent individual samples and the columns represent the 45 CpG sites (labeled with the CpG site unique ID and the gene associated with these sites). Euclidean clustering shows that samples from each tissue type are clustered together. The two paternally methylated CpG sites (in the paternally methylated GNAS/NESP55 DMR) are on the right of the heatmap and showed methylation patterns in uniparental tissues that are opposite to the other maternally methylated CpG sites. The samples are labeled white blood cell (WBC), placenta, androgenetic complete hydatidiform mole (AnCHM), and mature cystic ovarian teratoma (MCT). Note that each DMR is represented by one or several CpG sites and for detailed information about these CpG sites see Supplemental Table 1. (B) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent CpG methylation for five consecutive CpG sites within the maternally methylated KvDMR1 for each tissue. (C) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent CpG methylation for three consecutive CpG sites within the paternally methylated H19 DMR for each tissue. For B and C the bisulfite pyrosequencing data are derived from 15 blood samples, 10 placenta samples, three AnCHM samples, one paternal UPD11p15, and one MCT sample. (D) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent methylation for five consecutive CpG sites within the paternally methylated IG-DMR for each tissue. The bisulfite pyrosequencing data are derived from 15 blood samples, nine placenta samples, three AnCHM samples, one paternal UPD14 blood sample, one MCT sample, and one maternal UPD14 blood sample.











