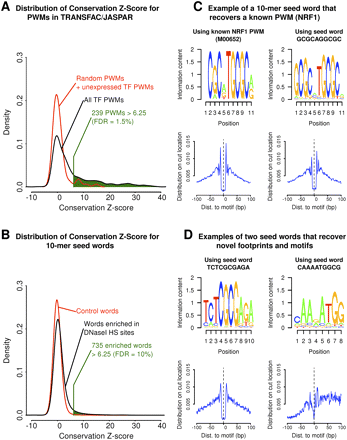
Application of the CENTIPEDE model across 756 known PWMs and 17,224 10-mers enriched in DNase I–sensitive regions. (Left panel) The distribution of the conservation Z-score for PWM motifs (A) and 10-mer derived motifs (B). (C) This panel shows that NRF1 binding sites are recovered from both the PWM and 10-mer-based approach. (D) Two examples that appear to be novel binding site motifs. For these motifs, locations inferred to be bound are strongly conserved, show a clear DNase I footprint, and show minimal overlap with known PWMs.











