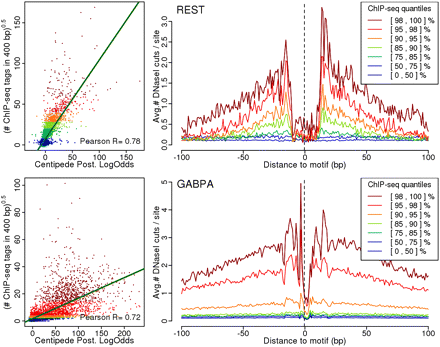
Footprint strength as a measure of TF occupancy. For REST and GABPA, respectively, we plot the posterior log odds from the CENTIPEDE model against the square-root transformed count of ChIP-seq reads in the 400-bp region surrounding each motif. Additionally, we plot the DNase I footprint for motif sites falling into seven quantile ranges of the ChIP-seq data. In each plot, the data are colored according to the quantile range in the ChIP-seq data. For both factors, there is a clear correlation between CENTIPEDE posteriors and the number of ChIP-seq reads. Furthermore, there is a clear gradient in the footprints going from the most pronounced footprint in the highest ChIP-seq quantiles to virtually no footprint in the lowest quantiles.











