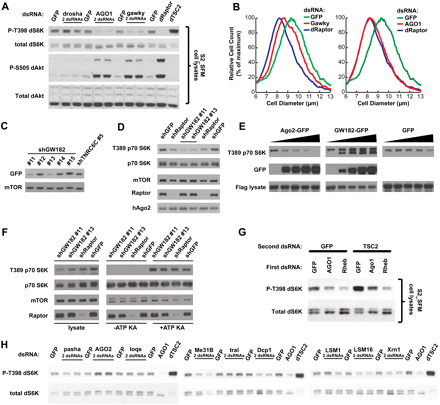
Validation and functional characterization of AGO1, gw, and related genes (cluster 3) in Drosophila and human cells. (A) Drosophila cells were transfected with the indicated dsRNAs, and lysates were analyzed by Western blotting. (B) Drosophila cells were transfected with dsRNAs targeting the indicated genes, and cell size distributions were obtained using a Coulter counter. Cell count is normalized to place the peaks of each distribution at equal heights; see also Supplemental Figure S2 for plots of absolute cell count as a function of cell diameter. (C) Human HeLa cells were transfected with a plasmid expressing a GFP-tagged GW182 and simultaneously infected with lentivirus expressing one of five GW182-targeting hairpins (#11–#15) or a hairpin targeting the related gene TNRC6C. Knockdown efficiency was determined by immunoblotting with an antibody recognizing GFP. (D) The shRNAs validated in C were used to knock down GW182 in HeLa cells. Lysates were prepared 3 d after infection and analyzed by Western blotting. The same shGFP and shRaptor samples on each side of the blot were loaded twice. (E) Human 293T cells were co-transfected with increasing amounts of AGO2-GFP, GW182-GFP, or pLJM1-GFP plasmids (0, 50, 100, 200, 400 ng) along with 2 ng of FLAG-S6K1-expressing plasmid. FLAG-S6K1 was purified, eluted off the Flag beads, and analyzed by Western blotting with an antibody against pT389. Lysates were also analyzed by immunoblotting. (F) mTORC1 was isolated through mTOR immunoprecipitations from HeLa cells 3 d after infection with shGW182 lentiviral constructs, and kinase assays were performed on S6K1 substrate. (G) Drosophila cells were incubated with the indicated dsRNAs in sequential order, with 2 d of incubation in the first dsRNA followed by 4 d of incubation in a mixture of the second and first dsRNA. (H) Drosophila cells were transfected with dsRNAs targeting the indicated regulators of RNA processing, and Western analysis revealed the relative effects on p-T398-dS6K of knockdown of these genes versus AGO1.











