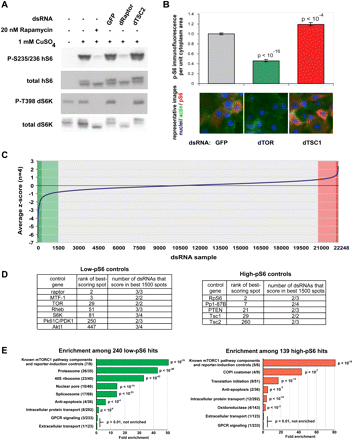
Genome-scale RNAi screen using a TORC1–S6K pathway immunofluorescence reporter identifies families of genes that regulate phospho-S6 levels. (A) The Drosophila S2R+ cell line was transfected to express the human S6 ribosomal protein under a metallothionine promoter, allowing induction by CuSO4 treatment. In a stable clone of these cells, called S6_S2R+, Western blotting demonstrates that phosphorylation of human S6 is sensitive to rapamycin as well as to dTORC1-pathway RNA1 (dRaptor and dTSC2); the reporter responds similarly to the endogenous Drosophila S6K, for which immunofluorescence-compatible antibodies are not available. (B) This cell line was transfected on microarrays with the indicated dsRNAs, and pS6 immunofluorescence per unit cytoplasm area was quantified with CellProfiler software (data shown as mean ± standard error, n = 16 spots). The dsRNA labeled “GFP” has no identical 19+mers to any transcripts in the genome, has no effect on TORC1 signaling, and is used throughout this work as a transfection control. (C) We screened, in quadruplicate, a genome-scale RNAi library containing 22,248 microarray spots targeting 13,618 Drosophila genes, for regulators of pS6 immunofluorescence. Candidate high-pS6 (red) and low-pS6 (green) hits were determined by assigning a Z-score to each spot on the array and combining scores for replicates. Candidate hit genes showed at least one dsRNA having a very strong phenotype (deeply shaded) or multiple dsRNAs having a moderately strong phenotype (lightly shaded); see the Supplemental Methods for more details. We found 240 candidate low-pS6 hits and 139 candidate high-pS6 hits. (D) The candidate hits included all the expected nonredundant components of the TORC1–S6K pathway, as elaborated in the text. (E) Genes were grouped by GO annotation (and into several ad hoc groups; see Methods), and gene groups were screened for fold enrichment among low-pS6 and high-pS6 candidate hits. For a given group, parentheses denote (number of group members scoring)/(number of group members in library). P-values are corrected for multiple comparisons (Benjamini and Hochberg 1995).











