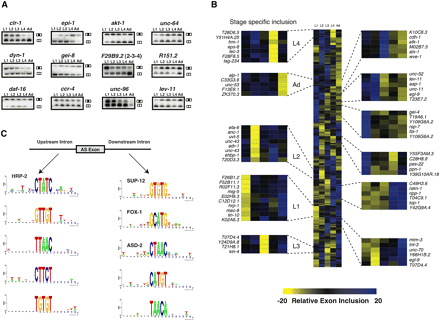
Identification of temporally regulated AS events. (A) Semiquantitative RT-PCR validation for a subset of regulated AS events across larvae (L1, L2, L3, and L4) to adult (Ad) stages. Primers were designed to amplify both isoforms, and in each case, the two possible isoforms (top bands, included isoform; bottom bands, excluded isoform) show changes in relative intensities across developmental stages. (B) AS exons that were identified to undergo temporal regulation across development of at least 20% were median centered and hierarchically clustered and visualized using Cluster and TreeView (Eisen et al. 1998). Blue boxes indicate higher exon inclusion, while yellow boxes indicate higher exon exclusion. We find exons that are excluded in specific stages (projected panels to the left) while we also notice shared patterns of relative exon inclusion between multiple stages for other subsets of AS events (projected panels to the right). (C) SeedSearcher was used to identify pentamer motifs that are statistically enriched in either the 50 nt upstream of or downstream from splice junctions that are differentially regulated during development. Examples of these enriched motifs are shown either for motifs located upstream of the AS exon (left) or downstream from the AS exon (right). Note that four of these motifs correspond to the published motifs of HRP-2, SUP-12, FOX-1 and ASD-2, and these are indicated.











