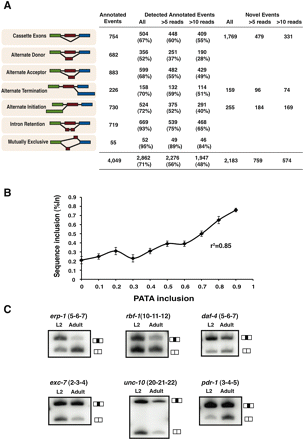
Identification of known and novel AS events. (A) Table displaying the proportion of different classes of AS events annotated in WormBase that are identified by our RNA-seq analysis with varying degrees of junction count support. Also listed in the last three columns are the number of unannotated AS events identified by our analysis and the number of reads supporting these splice junctions. (B) We compared the %In values for a set of 317 annotated cassette AS events from the true positive set that were detected both by RNA-seq analysis and by our microarray platform. For these AS events, RNA-seq derived %In values profiled across four developmental stages (L2, L3, L4, and adults) were compared to the PATA derived %In values in the same stage. The plot shows a high correlation between the two platforms indicating that both methods are reliable estimates of relative isoform usage. (C) RT-PCR validation of a subset of unannotated AS events in the larval (L2) and adult developmental stages. Primers were designed to anneal to sequences corresponding to neighboring constitutively spliced exons, amplifying two products: one representing isoforms including the alternative exon in transcripts (top bands in gel images) and the other one skipping the alternative exon (bottom bands).











