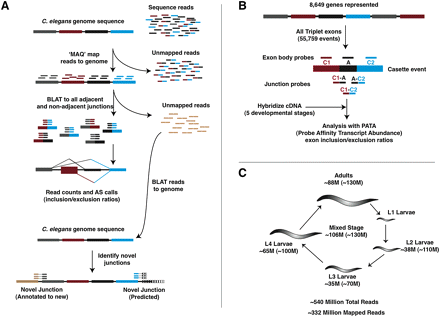
Outline of RNA-seq and microarray approaches for profiling AS in C. elegans. (A) For RNA-seq analysis, short sequence reads from the transcriptomes of animals isolated at multiple or specific developmental stages are first aligned to the genome and those that uniquely map in their entirety are used to identify exonic regions of transcripts. The remaining unmapped reads are then mapped against an exon junction database containing all possible combinations of splice junctions between annotated exons to identify and quantitatively measure AS events. Finally, any remaining unmapped reads are aligned to novel splice junctions that are not annotated or predicted. (B) Our microarray platform uses exon body (labeled C1, A, and C2 in diagram) and exon junction (labeled C1:A, A:C2, and C1:C2) probes to monitor cassette-type AS events, where the internal exon in a triplet of exons can be skipped in spliced mRNAs. We have included probes to monitor a total of 55,759 exon triplets in 8649 genes (∼50% of annotated genes), in addition to a set of 499 previously annotated cassette AS events. Probe sets corresponding to exon triplets with evidence of AS by our RNA-seq analysis are then combined with the PATA algorithm to generate quantitative predictions of relative isoform usage across the developmental stages analyzed. (C) Number of sequence reads uniquely mapping to the C. elegans genome by stage and sample. The values in parentheses indicate the total number of reads generated for each sample.











