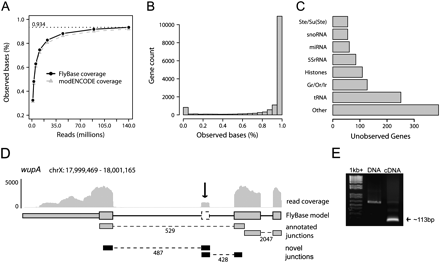
Deep RNA-seq covers 93% of the annotated D. melanogaster transcriptome. (A) 93.4% and 91.9% of the FlyBase and modENCODE annotated transcriptome is observed by pooled RNA-seq reads. Simulated read densities indicate that the current depth of sequencing approaches saturation. (B) Annotated transcripts are well covered by RNA-seq reads. More than 70% of annotated genes are covered for >95% of their length. (C) Most unobserved genes can be categorized into classes which are not expected to be observed due to size, genomic duplication, or lack of polyadenylation. (D) A novel exon is identified in wupA, which is supported by two novel junctions with 487 and 428 reads, respectively. (E) RT-PCR designed to validate the novel junction successfully amplifies an appropriately sized product from cDNA but not from genomic DNA.











