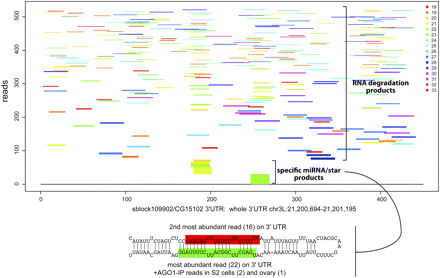
Example of a transitional miRNA locus, which exhibits signatures of both RNA degradation as well as Drosha/Dicer-1 processing across its precursor. Each read length has been plotted in a distinct color to emphasize the heterogeneity of cloned species mapping to the 3′ UTR of CG15102. The reads have been ordered on the y-axis with the most abundant individual species at the bottom. It can clearly be seen that a specific set of 21–22 nt reads are specifically made. These map to a typical pri-miRNA hairpin with a lower stem and a miRNA/miRNA* duplex region.











