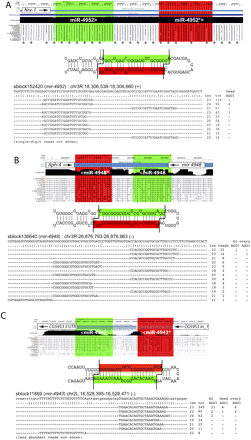
Examples of novel miRNAs generated from mRNAs. (A) A miRNA from the sense strand of the Nrx-1 coding region. This locus generates a specific miRNA/miRNA* duplex and exhibits some reads from head AGO1-IP data. Inspection of 12 species alignments indicates that the hairpin sequence evolves readily by codon wobbles, at a rate similar to the flanking nonhairpin codons. (B) A miRNA from the antisense strand of the Aph-4 coding region. In addition to specific miRNA/miRNA* duplex reads, this locus also generated a phased 5′ moR. (C) miRNA production from a primary-mRNA transcript in which the hairpin is produced from the pairing of intronic and exonic sequence of CG5953.











