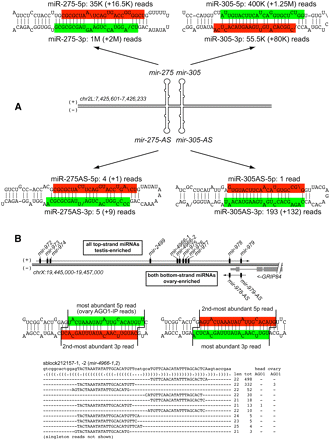
Examples of antisense transcription and processing across miRNA operons. (A) The top genomic strand of the mir-275/mir-305 locus is abundantly converted into mature miRNAs, but the bottom genomic strand also exhibits confident evidence for miRNA production across both miRNA hairpins. Primary numbers indicate reads matching precisely to the highlighted species; numbers in parentheses sum all other isomiRs matching that hairpin arm. (B) The distal end of the mir-972-979 cluster on the X chromosome overlaps Grip84, transcribed on the other strand. We detected confident miRNA production from the antisense strands of mir-979 and mir-978. This locus also bears a perfect tandem hairpin (sblock212157/mir-4966) that is subject to alternate Drosha and Dicer-1 cleavage to produce multiple 5′ isomiRs on both hairpin arms; multiple species were also detected in AGO1-IP libraries. The entire hairpin is duplicated; thus, all reads could map to either location.











