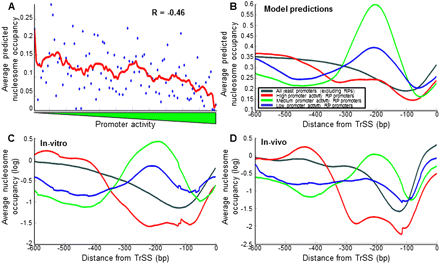
RP promoters with high promoter activities have a lower intrinsic affinity to nucleosomes. (A) For each RP promoter, shown is the lowest average nucleosome occupancy, predicted by a computational model of nucleosome sequence preferences (Kaplan et al. 2009), across any 10-bp region within 200 bp upstream of the translation start site (y-axis). RP promoters are plotted by their measured promoter activity (x-axis). Also shown is a moving average of consecutive RP promoters using a window of 11 promoters (red line). (B) For each group of RP promoters with high (red), intermediate (green), and low (blue) promoter activities, defined as in Figure 4, the average model-predicted (Kaplan et al. 2009) nucleosome occupancy across the group promoters is shown as a function of the distance from the translation start site. (C) Same as in B, but using measurements of nucleosome occupancy in vitro (Kaplan et al. 2009), in which nucleosome positions are dominated by nucleosome sequence preferences. (D) Same as in B, but using measurements of nucleosome occupancy in vivo (Kaplan et al. 2009) during growth in glucose-rich media.











