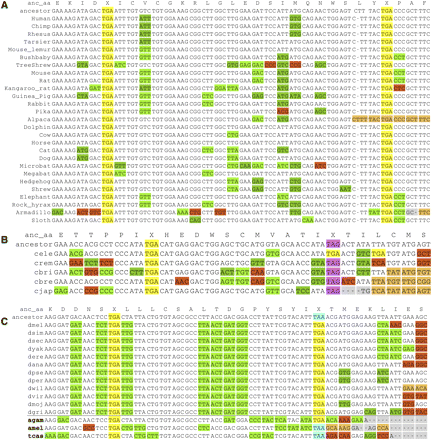
Examples of readthrough candidates in other species. (A) Alignment across 29 mammals for readthrough region in human gene SACM1L, one of four mammalian candidates. (B) Alignment across five worm species for the readthrough region in C. elegans gene C18B2.6, one of five nematode candidates. The stop codon context in all five is TGA-C and is perfectly conserved among Caenorhabditis species. (C) Alignment across 12 Drosophila and three other insect species, Anopheles gambiae (mosquito), Apis mellifera (honey bee), and Tribolium castaneum (red flour beetle), for the readthrough region of the D. melanogaster slo gene, one of 17 readthrough candidates conserved in mosquitoes, and one of four conserved across all 15 aligned insects. Although PhyloCSF cannot tell us whether the region is protein-coding in a particular subset of species, the large number of synonymous substitutions specifically in the other three insects, lack of non-synonymous substitutions and frameshifting indels, and perfectly conserved “leaky” TGA-C stop codon context suggest that readthrough also occurs in these other insects.











